|
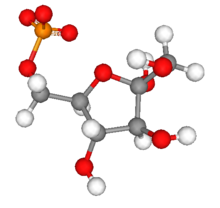
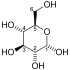
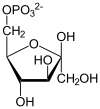
Fructose 6-phosphate
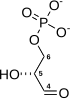
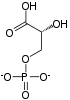
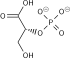
Fructose 6-phosphate (sometimes called the Neuberg ester) is a derivative of fructose, which has been phosphorylated at the 6-hydroxy group. It is one of several possible fructosephosphates. The β-D-form of this compound is very common in cells.[1][2] The great majority of glucose is converted to fructose 6-phosphate upon entering a cell. Fructose is predominantly converted to fructose 1-phosphate by fructokinase following cellular import. HistoryThe name Neuberg ester comes from the German biochemist Carl Neuberg. In 1918, he found that the compound (later identified as fructose 6-phosphate) was produced by mild acid hydrolysis of fructose 2,6-bisphosphate.[3] In glycolysisFructose 6-phosphate lies within the glycolysis metabolic pathway and is produced by isomerisation of glucose 6-phosphate. It is in turn further phosphorylated to fructose-1,6-bisphosphate.
Compound C00668 at KEGG Pathway Database. Enzyme 5.3.1.9 at KEGG Pathway Database. Compound C05345 at KEGG Pathway Database. Enzyme 2.7.1.11 at KEGG Pathway Database. Enzyme 3.1.3.11 at KEGG Pathway Database. Reaction [1] at KEGG Pathway Database. Compound C05378 at KEGG Pathway Database. Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
Glycolysis and Gluconeogenesis edit
See alsoReferences
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Portal di Ensiklopedia Dunia